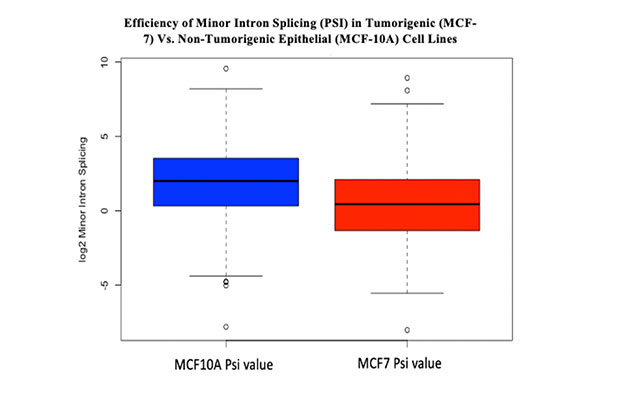
Minor introns are remarkably found in rare but a highly conserved sequences where there are only around 800 minor introns that function in cell cycle control, DNA damage repair, signal transduction and information relay contained within 20,000- 25,000 human genes. They provide a rapid response to cell stress serving as molecular switches that provide a rapid control of gene expression without the need of transcribing new pre-mRNA. These special introns and their mode of splicing are highly prevalent in cancer. Cancer is a disease where there is an abnormal cell growth that forms a mass resulting from a mutation in genes. These changes result from several environmental factors including radiation, chemicals (often called carcinogens), viruses and other infections and genetics. This project focuses on breast cancer which is a cancer that forms in breast cells. It is the most prevalent form of cancer in Qatar with an incident rate of 7.41 in females and 1.96 in both genders. This project is a computational analysis and of minor introns in breast cancer to better understand the regulation of those minor introns-containing genes in cancerous cells, their splicing pattern, their fragments produced and the factors that are possibly affecting any deviation observed from non-cancerous cell lines. Methods involved included using several databases and online tools such as Gene Ontology, gene cards and R studio to do the comparative studies. Two cell lines were used; the cancerous MCF7 and the non- cancerous MCF-10A. They are an excellent model to study because they are easily cultured (immortal) and well-characterized in terms of content and their expressed proteins.