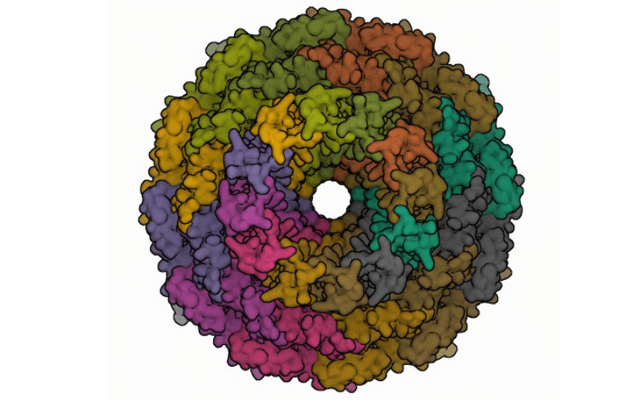Phages are viruses that have evolved to target one or several similarly related bacterial strains. As bacteria develop drug resistance, humans have turned to phages as an alternative form of antibacterial treatment. We need to identify phages that are effective at destroying hazardous bacteria. In this research, an unknown phage genome has been isolated and analyzed to learn about its characteristics and potentially identify the bacteria it targets and lyses. The process from reads files isolated in the lab and assembled using gene sequencing software. Afterward, careful analysis of the genome and the use of public genetic databases helped identify potential proteins within the genome. So far, 50,000 of 85,000 base pairs have been analyzed and their proteins have been identified 4 of which with their specific functions. The 4 proteins were: T4 Phage Tail Protein (used for attaching and penetrating the cell membrane), Thymidylate Synthase (used for nucleotide production), T7 Leading Strand DNA Polymerase (part of a larger protein unit designed to copy DNA effectively), and the Portal Protein (acts as a foundation in which to assemble the phage’s components into one cohesive unit). This phage shares many proteins with E. Coli phages, Salmonella Phages, as well as the hosts’ cells E. Coli, and Salmonella, leading to the conclusion that this phage must either be an E. Coli phage or a Salmonella Phage.